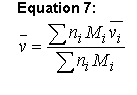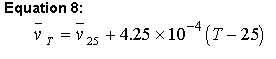Difference between revisions of "Corrected"
Bill Bashir (talk | contribs) (Created page with "The partial specific volume vbar is a measure of the change in the volume (in ml) of the solution per gram of solute. For typical proteins, any errors in vbar are magnified 3-fol...") |
(No difference)
|
Revision as of 14:47, 11 August 2011
The partial specific volume vbar is a measure of the change in the volume (in ml) of the solution per gram of solute. For typical proteins, any errors in vbar are magnified 3-fold in the determination of molecular weight (Ref. 9). Therefore, good estimates of are necessary. An excellent discussion of partial specific volumes, along with an extensive tabulation of values, may be found in the review by Durschlag. (Ref. 6) Several methods are available for the measurement of vbar, but these typically require relatively high concentrations of solute, may require specialized equipment and definitely require careful analytical techniques for their success (ref. 6 40).
An alternative to measuring vbar is to estimate its value based on the protein's composition. Comparison of measured and estimated vbars indicate that the error typically is less than 1%. (Ref. 40) Somewhat larger errors are observed for proteins that have an unusual amino acid composition, proteins that are conjugated with carbohydrate, proteins that exhibit preferential hydration or proteins that have a non-globular structure. (Ref. 6 40 41) Even under these circumstances, the error is typically less than 3% if the correct composition is used. (Ref. 6 40)
The partial specific volume usually is estimated using the method of Cohn and Edsall (Ref. 4 5 6 40):
Equation 7:
where n is the number of moles of the ith component and M is its molecular weight. Values of vbari for the amino acids and several protein-associated components stored in the phyconst database. Note that traditionally the M values used are the residue weights, i.e. they have been adjusted to reflect the loss of H2O associated with the formation of the amide linkage. This means that this formula essentially ignores both the molecular weight and the molar volume of the amino-terminal proton and the carboxy-terminal hydroxyl (and that is the implementation used in Sednterp).
Sednterp uses the values in the database to calculate at 25 degrees Celsius and then uses equation 8 to adjust this value over a temperature range 4-45 of oC using (Ref. 6):
Equation 8:
(note that this equation is incorrect in ref. 59)
There is no computational method to correct for the effects of pH. However, it has been observed that varies only slightly with pH unless denaturation occurs, at which point decreases by about 3%. This decrease in with pH parallels other changes in the protein structure (e.g. unfolding as monitored by viscosity or by circular dichroism).
Buffer composition and preferential solvation also can effect (Ref. 6), but these are usually neglected except for denaturing buffers.
All of the partial specific volumes in the phyconst database have been determined for unconjugated molecules. For a few non-polar ligands, this approximation has been shown to be reasonable, however, caution should be exercised when calculating partial specific volumes for lipoproteins. (Ref. 35) Likewise, greater uncertainty results from the use of determined for unconjugated carbohydrates. (Ref. 41)